TLDR
- Google DeepMind CEO Demis Hassabis and John Jumper, senior research scientist, were awarded the Nobel Prize in Chemistry for developing AlphaFold2, which predicted the structure of over 200 million proteins. They solved a 50-year-old biochemistry challenge.
- Hassabis and Jumper share the Nobel with David Baker from the University of Washington, who has created entirely new proteins.
- Proteins form the building blocks of life. Knowing their structure can help researchers develop drugs and treatment for diseases stemming from malfunctioning proteins, such as Alzheimer’s.
What a week for artificial intelligence. A day after the Royal Swedish Academy of Sciences decided to give the Nobel Prize to two AI pioneers – despite no award category for computer science – the judges have done it again.
This time, the Nobel Prize in Chemistry goes to Google DeepMind CEO Demis Hassabis and senior research scientist John Jumper for developing AlphaFold2, an AI model that predicted the structure of almost all known proteins, solving a 50-year-old biochemistry problem. They share the Nobel with David Baker, from the University of Washington, who has created entirely new proteins.
“Both of these discoveries open up vast possibilities,” said Heiner Linke, chair of the Nobel Committee for Chemistry, in a statement.
Proteins are important because they serve a wide variety of critical functions in the body, including building and repairing tissues, enzymatic functions, transporting and storing molecules and nutrients, regulating hormones, defending the body (antibodies are proteins), and as an energy source.
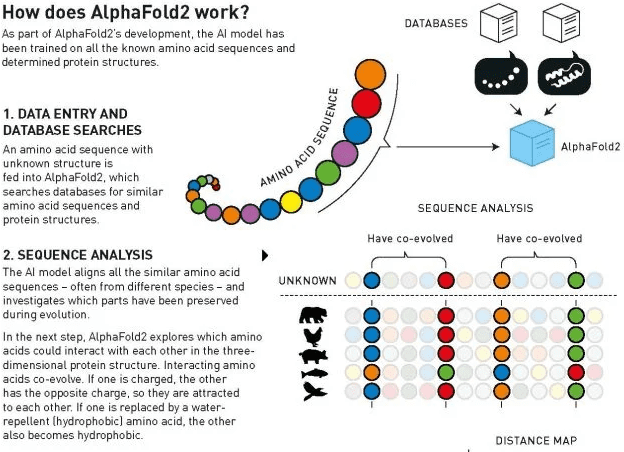
It’s important to know a protein’s structure, since this determines its role in the body. Scientists can then study diseases caused by malfunctioning proteins, design drugs that target specific proteins, develop new proteins with customized functions, and treat diseases such as Alzheimer’s. But finding and predicting the structure can take years and hundreds of thousands of dollars. Moreover, there are over 200 million known proteins, most of which don’t have solved structures.
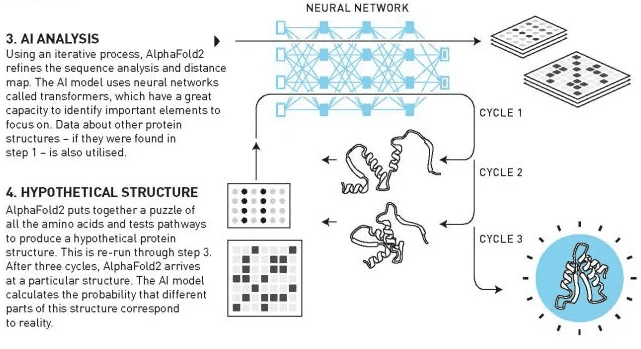
AlphaFold2 was able to predict the 3D structures of “virtually all” 200 million proteins from a sequence of amino acids. Hassabis and Jumper, who co-led the team, trained the AI model on a vast dataset of all known protein structures and amino acid sequences, using the new AI architectures.
“It is considered as the greatest contribution of artificial intelligence to the scientific field and one of the most important scientific breakthroughs made by mankind in the 21st century,” according to a paper by Sichuan University researchers, published last year in Nature.
Listen to an AI-generated podcast discussing the DeepMind academic paper on AlphaFold2: